Gene regulation in Bacteroides fragilis
How does Bacteroides fragilis adapt to the dynamic environment of the human gut? A newly published review paper by Daniel Ryan (IBB PAS) explores this question.
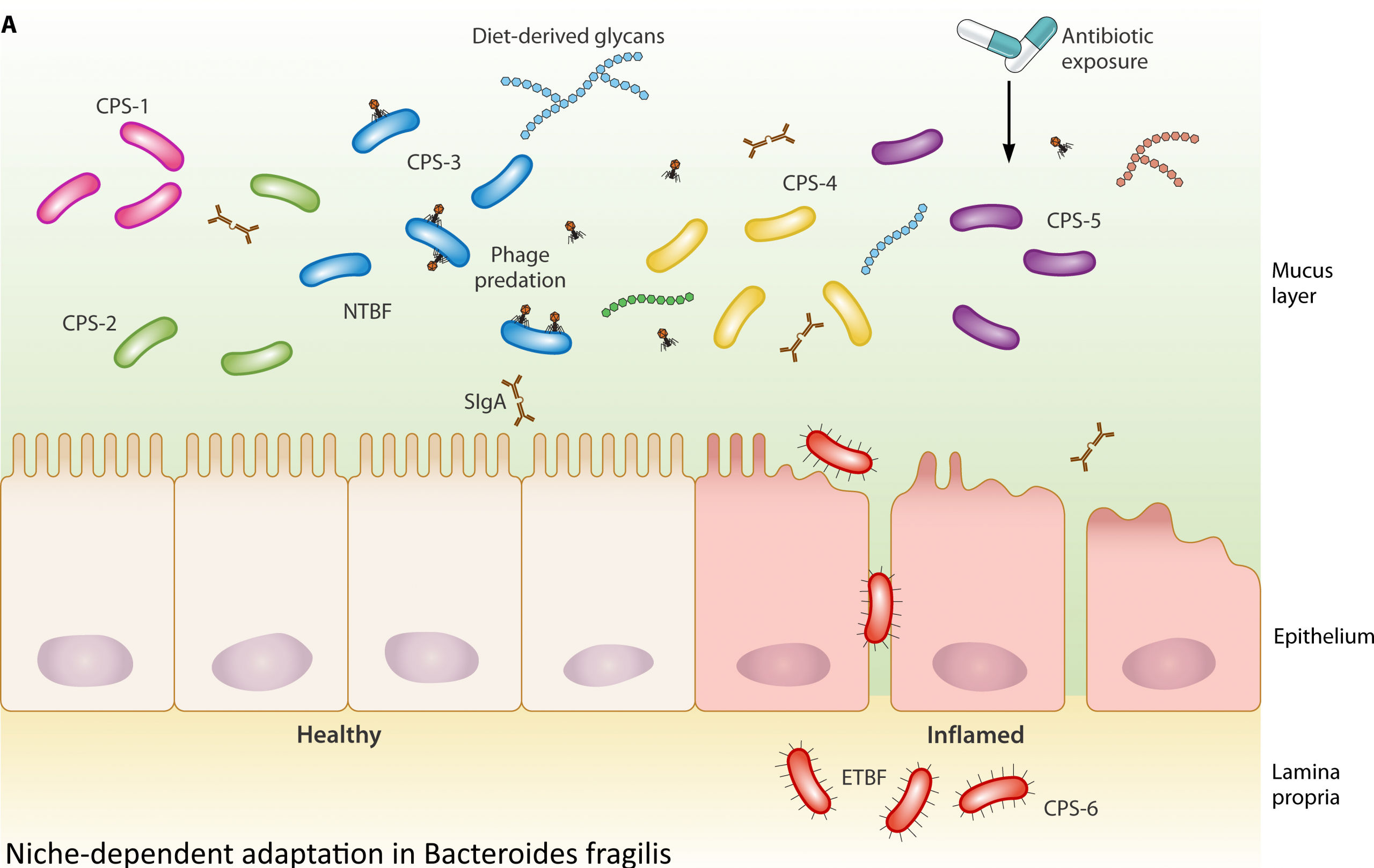
Bacteroides fragilis employs a modular gene-regulatory network strategy to respond to its environment. Rather than relying on global regulators, it coordinates survival via modular switches such as DNA inversions, phase-variable sigma factors, hybrid two-component systems, and small RNAs – mechanisms that collectively balance commensal persistence with pathogenic potential. The article positions B. fragilis as a model for studying flexible transcriptional control in complex microbial ecosystems.
Read the full paper in Microbiology and Molecular Biology Reviews: https://journals.asm.org/doi/10.1128/mmbr.00225-25