The dynamics of mRNA deadenylation
Scientists from IBB PAS, IIMCB, Aarhus University and University of Turku analyzed the dynamics of deadenylation of polyadenosine (pA) tails of mRNA in the yeast Saccharomyces cerevisiae under basal and stress conditions.
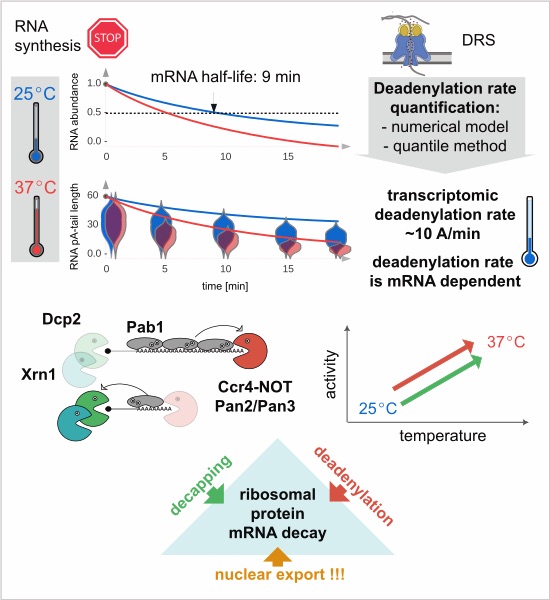
pA tails are an evolutionarily conserved modification of the 3′ end of mRNA, which is required for export of the transcript from the nucleus to the cytoplasm and its translation. The adopted models show that the successive shortening of the pA tail determines the duration of the transcript in the cytoplasm. Transcripts that have undergone significant deadenylation are decapped and degraded from the 5′ end by the exonuclease Xrn1.
In their work, scientists from IBB PAS and IIMCB created a model of transcriptome deadenylation based on a modified gamma distribution, thanks to which they determined that the deadenylation rate is 10 adenosines/min. In addition, they proposed a simplified method for calculating deadenylation rates based on changes in the distribution quantile values. These tools enabled visualization of the correlation between the rate of deadenylation and successive mRNA degradation and showed that both processes change in response to stress conditions. Interestingly, about 120 mRNAs encoding protein subunits of the ribosome, which by mass represent 40% of the transcriptome, are an exception and, in addition to deadenylation, require efficient ongoing export from the nucleus for degradation.
The full version of the article entitled “Modeling of mRNA deadenylation rates reveal a complex relationship between mRNA deadenylation and decay”, published in the EMBO Journal, is available here: https://www.embopress.org/doi/full/10.1038/s44318-024-00258-3.